ONGOING PROJECTS AND MAIN LINES OF RESEARCH
Plant evolution and diversification associated to mating system transitions
Especiación ecológica en plantas dirigidas por interacciones ecológicas en cascada: una aproximación experimental
(2023-2024)
Funded by the Spanish Association for Terrestrial Ecology. PI: Ana García-Muñoz
(2023-2024)
Funded by the Spanish Association for Terrestrial Ecology. PI: Ana García-Muñoz
|
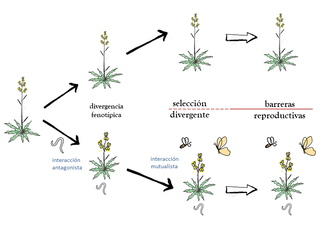
Las interacciones ecológicas conforman uno de los principales motores de evolución gracias a que generan presiones selectivas importantes. Estas presiones selectivas pueden ser divergentes y desembocar en la aparición de barreras reproductivas, relación causal que se conoce como especiación ecológica. Sin embargo, no es frecuente que este proceso comience con la aparición de interacciones de tipo antagonista (infección por nematodo) que modifiquen o faciliten interacciones de tipo mutualista (polinización). Unas y otras interacciones modifican las presiones selectivas haciéndolas divergentes entre plantas infectadas y no infectadas, mientras que los polinizadores además de ejercer una presión selectiva también intervienen en la aparición de barreras reproductivas. En poblaciones de Erysimum repandum hemos observado variación fenotípica entre individuos afectados por nematodos respecto a individuos sanos. En la presente propuesta pretendo explorar experimentalmente: 1) cómo afecta la interacción con nematodos al fenotipo de E. repandum y a la integración fenotípica de esta especie; 2) Evaluar las presiones selectivas divergentes ejercidas por los polinizadores sobre las plantas experimentales en las poblaciones naturales; 3) cuantificar las barreras reproductivas que estos polinizadores generan. Todo esto nos ayudaría a comprender mejor los mecanismos que subyacen y controlan los fenómenos de especiación ecológica, motor de la aparición de biodiversidad.
|
Understanding evolutionary transitions in plant mating systems: the “forbidden” path from selfing to outcrossing species (OUTevolution)
PID2019-111294GB-I00 (2020-2024)
Funded by the Spanish Ministery of Science, Innovation and Universities. PI: Mohamed Abdelaziz
PID2019-111294GB-I00 (2020-2024)
Funded by the Spanish Ministery of Science, Innovation and Universities. PI: Mohamed Abdelaziz
|
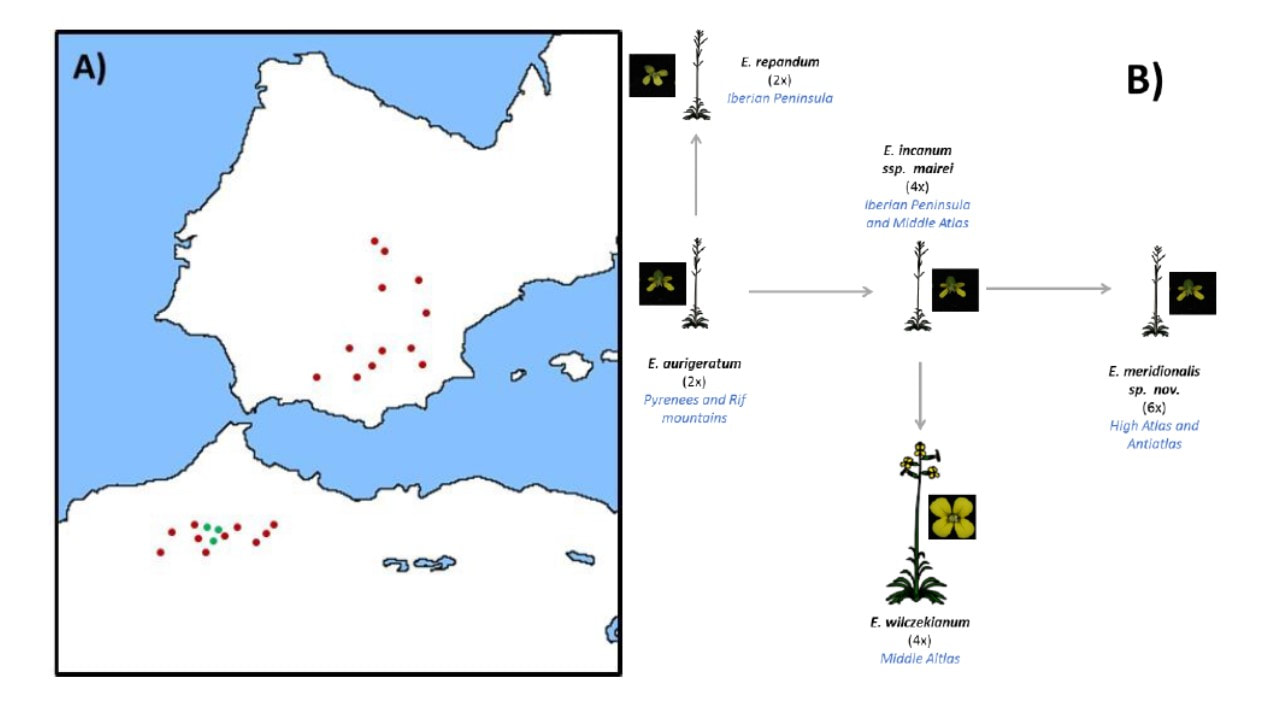
Since Darwin the shift from outcrossing to self-fertilization has been recognized as one of the most common evolutionary transitions in flowering plants. However, the lack of examples for transitions from self-fertilization to outcrossing in nature together with the population genetic theoretical framework, has made us consider it as a forbidden evolutionary path. Recently we found in the Erysimum incanum species complex solid evidences supporting that evolutionary transition to outcrossing in a selfing clade is possible in nature, giving us the opportunity to explore for the first time the mechanism, processes and consequences of this unknown evolutionary path. The current project (OUTevolution) aims to explore: (i) the genetic mechanism underlying this transition, using massive sequencing based technique and synthetic plants; (ii) the main ecological and genetic processes stabilizing the rise of new morphs and reproductive strategies in a selfing population, such as the pollinator interaction and the heterozygote advantage, respectively; (iii) and finally, the consequences of this evolutionary transition on the population structure in short term, the genome diversity in middle term and the genetic architecture in long term. Being the first research project exploring this kind of evolutionary transition, the results here obtained could represent a paradigm shift in our conceptual approach to speciation and diversification in plants. The project will shed light on the evolutionary rescue of outcrossing strategy in a selfing clade by using novel experiments reproducing artificially the evolution. OUTevolution will be also the first opportunity to explore the consequences of evolutionary transition to outcrossing on the population structure, genome diversity and genetic architecture. In addition, this project will point out the need to develop solid theoretical frameworks and methodological approaches to explore the speciation events in nature and the evolutionary transition associated to them, as responsible of the biological diversity that we can observe in nature, from a new point of view.
|
Mechanims of speciation in plants associated to mating system transitions (TransSpeciation)
CGL2014-59886-JIN (2015-2018)
Funded by the Spanish Ministery of Economy and Competitiveness. PI: Mohamed Abdelaziz
CGL2014-59886-JIN (2015-2018)
Funded by the Spanish Ministery of Economy and Competitiveness. PI: Mohamed Abdelaziz
|
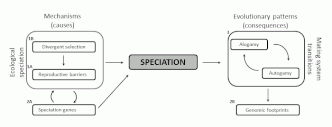
The speciation process triggers the emergence of new species and its study helps not only with understanding evolutionary questions but also with a better management of the natural biodiversity. The causes and consequences of speciation have drawn the attention of scientists from the beginnings of the evolutionary biology. However, while the ecological mechanisms and consequences of speciation were extensively explored in many organisms during decades, it was not until recent years, with the vertiginous development and increasing affordability of new sequencing technologies, when integrated approximations to the study of speciation, including its genomic aspects, have become increasingly feasible. The causes of speciation lie in a continuous interaction between ecological and genetic mechanisms. The importance of natural selection for creating barriers to gene flow between diverging taxa (ecological speciation) has been recognised. However, there are only few examples in which the identification of the selective pressures and reproductive barriers driving speciation were accompanied by the identification of genes underlying the speciation process. One of this consequences is the evolutionary transitions between diferent mating systems strategies in plants. In this project, we study a close related group of taxa included the incanum complex of species belonging to the genus Erysimum L. Although the species in the complex share many phenotypic and habit similarities, they strongly differ in floral sizes and pollinator assemblages. A multidisciplinary approach including the evaluation of the reproductive barriers and the study of the selective pressures driving them, together with the identification of the genetic mechanisms underlying these species divergence conform the main objectives of this project, together with the study of the evolutionary consequences of these speciation events.
|
Drivers of biodiversity changes in the global change era
Monitorization of Endemic ENdangered Ecosystems Registering genomic VAriation through time (MEENERVA)
PID2022-139405OB-I00 (2023-2027)
Funded by the Spanish Ministery for Science and Innovation. PI: A. Jesús Muñoz-Pajares.
PID2022-139405OB-I00 (2023-2027)
Funded by the Spanish Ministery for Science and Innovation. PI: A. Jesús Muñoz-Pajares.
|
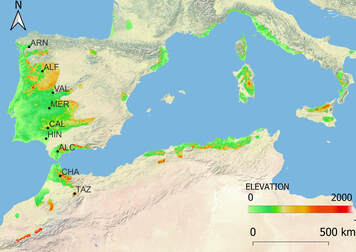
Climate change is affecting every ecosystem in the world but mountain summits are particularly threatened. In this project we will study genetic diversity trends of plant species from the Spanish Sierra Nevada, one of the most important plant biodiversity hotspots in Europe. The general objective of the MEENERVA project is to demonstrate that genomics can become a useful and inexpensive tool to describe and monitor biodiversity. For that, we will focus on ten taxa to develop different tasks that can be grouped into five objectives:
1) Delimiting taxa. The first step to quantify biodiversity is to characterise the taxonomic status of the taxa to be studied. Our study system includes one taxon showing unresolved taxonomic issues and one taxon showing potential introgression after a secondary contact with another close species. 2) Exploring current patterns of genetic variation. We will quantify the genetic diversity of every taxa across its entire distribution range in Sierra Nevada. Our study system includes one taxon lacking any close reference genome and we will produce a draft reference. 3) Quantifying past biodiversity loss during the last half-century. We will compare genetic diversity estimated from herbaria individuals collected more than 50 years ago from different locations with current individuals from the same populations. To compare patterns among taxa with different conservation status, our sampling will include no-threatened, endangered and critically endangered taxa. 4) Developing resources for monitoring biodiversity. We will develop reference databases and genetic diversity barcodes to address similar questions with much lower sequencing efforts in the future. 5) Conserving current biodiversity of Sierra Nevada taxa. To preserve the existing biodiversity of the studied taxa, we will collect seeds to be conserved in collaboration with the Herbaria of the Universities of Granada and Lisbon. Overall, the MEENERVA project will contribute to achieve the objectives of the Spanish Strategic Line Exploration, analysis and future of the biodiversity by producingr resources and information to develop appropriate conservation actions and avoid taxa extinction. |
Analysis of the INTERactive effects of multiple GLOBal change drivers on Mediterranean forests (INTERGLOB)
CNS2022-135560 (2023-2025)
Funded by the Spanish Ministery of Science and Innovation. PI: Luis Matías
CNS2022-135560 (2023-2025)
Funded by the Spanish Ministery of Science and Innovation. PI: Luis Matías
|
The increase in the frequency and intensity of drought events registered during the las decades originated by changes in climate, together with the impact of other global change drivers, is inducing a generalised decrease in tree health and vigour, finally causing massive forest mortality events across the globe. This dieback has an important impact on oak species, especially in the Mediterranean region, resulting in the so called “la seca” phenomenon. However, several global change drivers other than climate can simultaneously act over an ecosystem, inducing different impacts than the isolated effect of each of them. In this project, we will analyse the response of an ecologically and economically important ecosystem as the mixed cork oak forest to the combined effect of different global change drivers (changes in climate, invasive pathogens and atmospheric nutrient depositions). By means of a combination of observational and experimental approaches through environmental gradients, we will reach the following objectives: 1) To analyse the interactions between different global change drivers and their impacts on tree species; and 2) To evaluate the interactive impacts of different global change drivers on the ecosystem services that Mediterranean forests provide. The combination of milti-scale (from populations to individuals) and multi-disciplinary (joining advances in dendroecology, dendrochemistry, pathology and biogeochemistry) approaches will allow the generation of highly relevant knowledge from scientific and applied perspectives, as is the identification of especially sensitive areas to global change or the impacts over the ecosystem services that these forests provide. The results of this proposal will be of interest for the conservation of these important ecosystems, and will increase our knowledge about the interactive effect of different global change drivers.
|
Analysis of the causes and ecological consequences of forest dieback in response to global change (DECAFOR)
US-1380871 (2022-2023)
Funded by the Andalusian Government and University of Sevilla. PI: Luis Matías
US-1380871 (2022-2023)
Funded by the Andalusian Government and University of Sevilla. PI: Luis Matías
|
The increase in the frequency and intensity of droughts registered during the last decades due to climate change, together with other drivers of global change, are subjecting many species to strong adaptive pressures with negative impacts on forest dynamics, causing massive events of tree loss of vigor, defoliation and death around the world. This phenomenon of forest decay has an important impact among species of the Quercus genus, and especially in the Mediterranean Region, giving rise to the phenomenon commonly known as “la seca”. In this project, we will analyze the response of an important type of forest from an ecological and economic perspective as the mixed cork oak forest, to different drivers of global change and their implication in the alarming mortality rates registered in these species. Through a combination of observational, experimental and modeling approaches across a wide gradient of environmental conditions, we will try to achieve the following objectives: 1) To identify the early signals of decay in species of the Quercus genus; 2) To analyze the interactions between different drivers of global change; and 3) To model the response of mixed oak forests to expected changes in climate. The combination of multi-scale (from populations to individuals) and multi-disciplinary approaches (combining the latest advances in dendrochronology, ecophysiology and modeling) will allow the generation of a large amount of information of high scientific and applied relevance, such as the identification of especially sensitive areas to climate change and the mechanisms underlying dieback processes. The results obtained with this project will be relevant for the conservation of these important species, at the same time that it will provide knowledge about the factors and mechanisms that induce tree mortality.
|
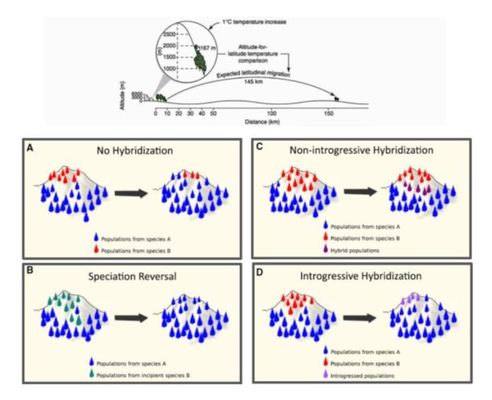
Hybridization as driver of biodiversity changes in the global change era: an experimental and predictive perspective of the phenomenon (globalHybrids)
SPIP2017-02415 (2020-2023)
Funded by the Autonomous Organism for National Parks. PI: Mohamed Abdelaziz
SPIP2017-02415 (2020-2023)
Funded by the Autonomous Organism for National Parks. PI: Mohamed Abdelaziz
|
The species hybridization is a phenomenon which consequences can go from the collapse and loss of diversity because of genetic introgression events, to the rise of this diversity because of hybrid speciation. The anthropic effects, including the global change, are promoting the secondary contact and hybridization between diverging taxa, especially in protected areas. That happens because different species (plants in particular) have not completed the development of reproductive barriers because of evolutionary divergence, and they are getting together because of changes in their altitudinal distribution. Understanding and predicting the consequences of these hybridization events seems crucial for any future conservation strategy and biodiversity management policy, more particularly in those areas with special conservation interest. In the current project we aim to study the ecological and genetic mechanisms promoting or preventing the past, present and future hybridization. For this, we think that an appropriated system are the different contact zones between Erysimum species (Brassicacea) in more than a half of the Spanish National Parks. Using this framework, we will genetic and ecologically characterize the different contact zones of these species of Erysimum. We will quantify the reproductive barriers between the contacting species and their hybrids. We also will explore the positive and negative dense-dependence processes using an experimental approach. And we will value how these processes are affected by the future global change predictions. Finally, we aim to use all that information to implement it in a predictive model which could be used in other systems with potential hybridization because of the global change, influencing on the biodiversity patterns of protected areas, identifying priorities for conservations policies.
|
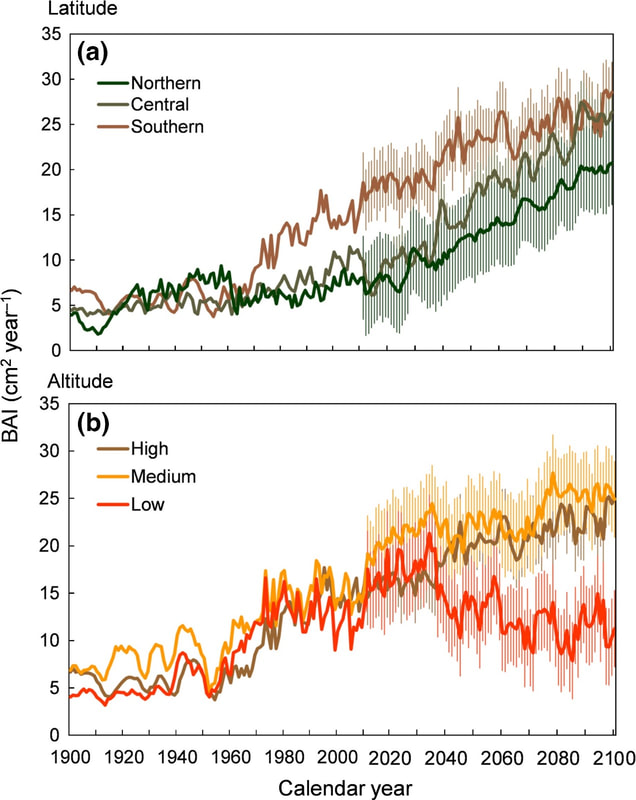
Mechanisms implied on the adaptation of Mediterranean forests to climate change (MEADCLIM)
PID2019-108288R (2020-2022)
Funded by the Spanish Ministery of Science and Competitiveness. PI: Luis Matías
PID2019-108288R (2020-2022)
Funded by the Spanish Ministery of Science and Competitiveness. PI: Luis Matías
|
The changes in climate registered during the last decades have the potential to modify the phenology, growth and biotic interactions for many plant species at global scale. However, the longer exposure of populations closer to the equatorial distribution limit to more extreme conditions might have induced the appearance of some adaptations related to increased environmental tolerance, as e.g. resistance to water stress, increasing the survival probabilities under a global-change scenario. In this project, we will use cork oak (Quercus suber L.), a species with high ecological and economic impact currently suffering a declining process, as a model organism. By means of a combination of observational, experimental and modelling approaches along the complete latitudinal distribution of the focal species, we will reach the following objectives: 1) To quantify and to model secondary growth and water use efficiency in natural populations along the species’ latitudinal distribution; 2) to determine the morphological traits and the physiological mechanisms implied on the response to drought and their tolerance thresholds; and 3) to identify the genetic mechanisms implied on drought resistance using transcriptomic and genomic techniques. The combination of multi-scale (from populations to individuals and genes) and multi-disciplinary (joining the latest advances in dendrochronology, modelling, ecophysiology, functional ecology, transcriptomics and genomics) approaches will allow the generation of a vast amount of information of high relevance from scientific and applied perspectives and the underlying mechanisms to acclimation and adaptation processes.
|
Drought adaptations in cork oak trees in a changing world (DRACO)
LRG-5603-6647 (2016-2018)
Funded by the British Ecological Society (BES). PI: Luis Matías
LRG-5603-6647 (2016-2018)
Funded by the British Ecological Society (BES). PI: Luis Matías
|
Recent changes in climate are altering the performance, growth and distribution of species at global scale. However, populations located in the most drought prone areas of a species distribution can harbour genetic adaptations that improve their survival probability under climate change. In this study, I will focus on Cork oak (Quercus suber L.), a species with high economic and ecological importance but one showing population decline during the last decade as a consequence of global change. Based on field investigations I will determine the changes in population dynamics that are currently operating in response to changes in climate across the complete distribution of the species (covering Morocco, Spain and Portugal). In parallel, I will examine specific changes in physiology, morphology and demography of seedlings from the wetter and more drought-prone edges of the species distribution in response to experimentally imposed climate change under controlled conditions. The information gained in this project will be key for the conservation of this imperilled and important tree species and provide vital information on how species adapt to extreme environments.
|
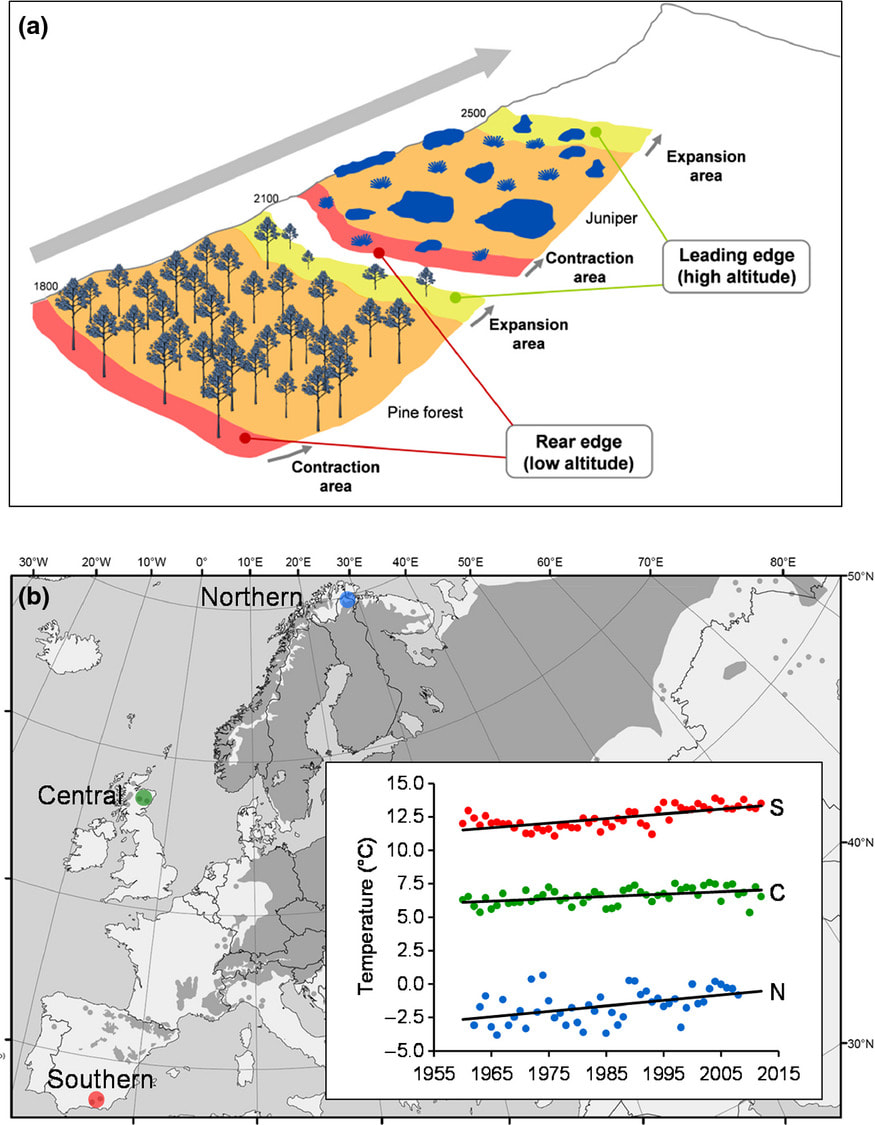
Can altitudinal data predict latitudinal responses of plants to climate change? (ALT-LAT-RANGE)
FP7-PEOPLE-2011-IEF-300825 (2012-2014)
funded by the European Union through a FP-7 Marie Curie IEF Fellowship. Co-PIs: Luis Matías & Alistair Jump.
FP7-PEOPLE-2011-IEF-300825 (2012-2014)
funded by the European Union through a FP-7 Marie Curie IEF Fellowship. Co-PIs: Luis Matías & Alistair Jump.
|
The distributional limits of species are frequently strongly determined by climate. Rapid changes in climate are disrupting environmental conditions throughout species ranges, with the most dramatic effects predicted to occur at the range edge. Changes in individual growth, survival and reproduction lead to population expansion or decline and induce range shifts, both at altitudinal and latitudinal limits of the distribution of a species. In this study, we are assessing the capacity for population expansion in altitude and latitude as a consequence of climatic change and evaluating the likely range contraction at species lowermost/southernmost distribution limits. We also aim to analyse discrepancies and similarities between altitudinal and latitudinal range shifts, given that range shifts are often reported in mountain areas but less frequently in the lowlands.
This research combines controlled environment experimental manipulation of growing conditions in of Scots pine (Pinus sylvestris) populations from the geographic limits of the species with field-based assessment of the performance of Scots pine and common juniper (Juniperus communis) across their altitudinal and geographical ranges. Controlled environment study is assessing ecological and physiological characters to look at emergence, morphology, growth and drought susceptibility. Filed-based research is looking at growth rate and its interannual variability, together with fruit production, establishment and the age structure of individuals within natural populations. We are also looking at biotic interactions through assessment of herbivory damage across the range of these species. Overall, this work will allow us to understand the impacts of current changes in climate on population performance in these species and the implications for their future distribution. |
Science to improve crops
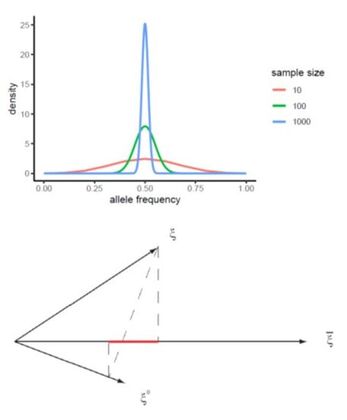
Methods, computational tools and empirical studies focused to the detection of the genomic signals due to selection by domestication (DOMSELEFFECTS).
PID2020-119255GB-I00 (2021-2024)
Funded by the Ministery of Science and Innovation (MICINN). PI: Sebastián E. Ramos Onsins
PID2020-119255GB-I00 (2021-2024)
Funded by the Ministery of Science and Innovation (MICINN). PI: Sebastián E. Ramos Onsins
|
This proposal aims to increase the understanding of the main mechanisms that drive the process of adaptation to fast (environmental) changes, such as domestication. The domestication process is an excellent model to study the genomic effects of selection because (i) there are species with domestic and wild populations to compare; (ii) we know that there has been selection on the domestic populations, since they have been progressively modified by human interest and (iii) the modification of the domesticated species has been rapid in terms of evolutionary time.
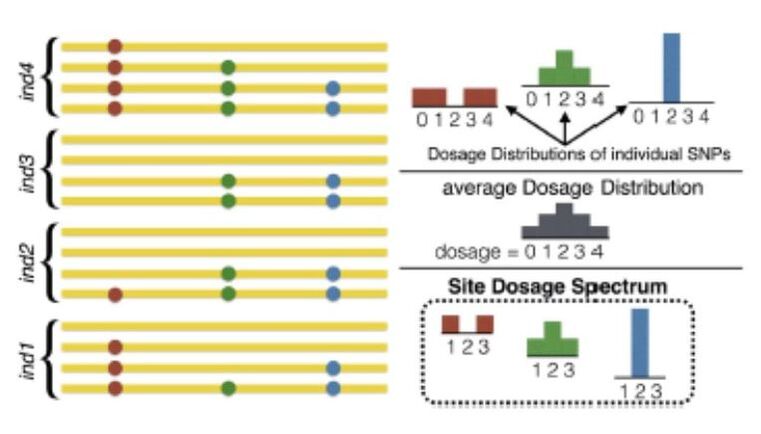
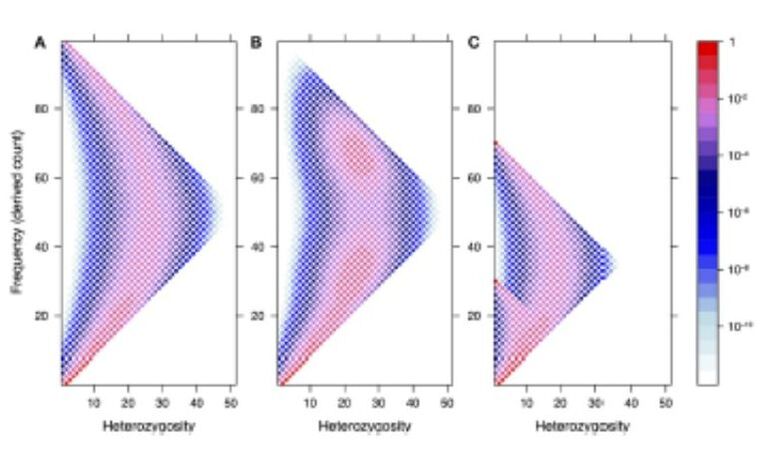
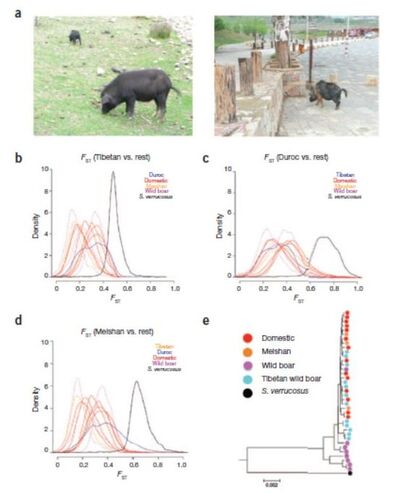
This proposal is a continuation of the previously funded projects (CGL2009-09346, AGL2013-41834, AGL2016-78709-R), in which the effects of selection will be explored through the inference of the distribution of selective effects in the genome, including the still under- explored polyploid species, or through a new proposed method that combines genotype-phenotype association with population genetic statistics. An interdisciplinary strategy has been adopted for this proposal: (i) development of theoretical and statistical methodologies; (ii) apply these methods and others available for the study of public empirical data (previously generated by our group and from public databases) to understand the process of adaptation of domestic populations, and (iii) transference of these methodologies to software tools to make them available for general public and researchers. The different strategies (methods, software, analysis) are inter-related and will be developed coordinately. Additionally, the software developed in this project, together with other tools that we have previously developed, will be considered by a Biotechnology company to be included in their analysis pipeline. Specifically, the methodologies to be developed are three: (i) the development of a Bayesian method to detect the sample frequency of SNPs from pools and polyploids with the objective of obtaining the unbiased site frequency spectrum; (ii) the development of an algorithm to estimate the distribution of selective effects in autopolyploid species and (iii) the development of a new statistical method to detect the effect of strong selective events associated with a given phenotypic trait, based on the linkage disequilibrium signal caused by selective sweeps. Computational code and the software applications will be developed at the same time than methods to validate them and to be used in empirical studies. These tools and others available will be used for the study of the differences of selective effect between domestic and wild populations, focusing on porcine and bovine for diploid species, and on potato for autotetraploid species. |
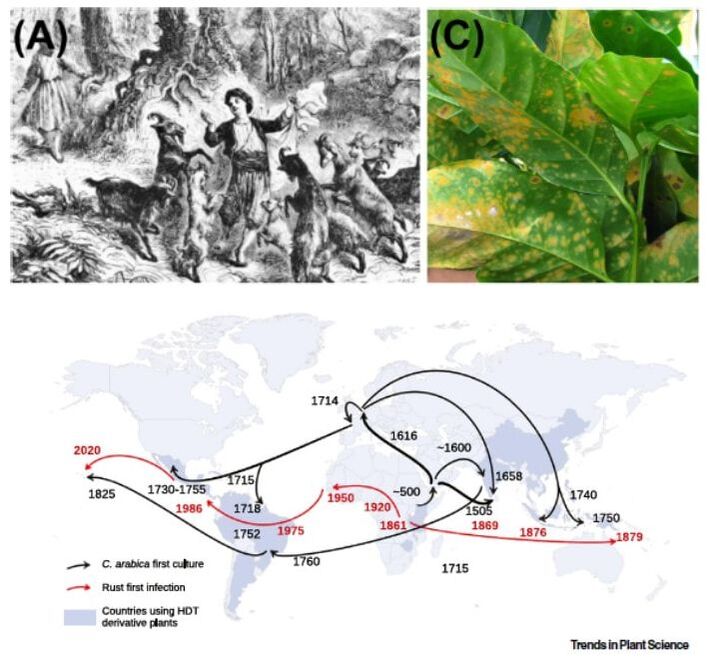
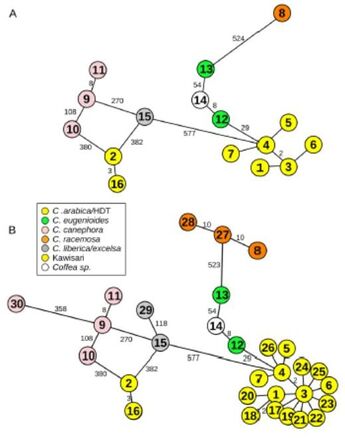
Unraveling Timor hybrid resistance to coffee rust by genomic approaches (HDT-Coffee)
PTDC/ASP-PLA/32429/2017 (2018-2021)
Funded by Portuguese Foundation for Sciences and Technology. PI: A. Jesús Muñoz-Pajares
PTDC/ASP-PLA/32429/2017 (2018-2021)
Funded by Portuguese Foundation for Sciences and Technology. PI: A. Jesús Muñoz-Pajares
|
Coffee is the most important agricultural product in international trade, with coffee leaf rust being the most economically impacting disease in the world. Portugal, through CIFC, was instrumental in the preservation and dissemination of the natural sources of rust resistance found in a spontaneous hybrid called ?Híbrido de Timor? (HDT), which was ultimately used during the production of most of the world-wide modern improved varieties. However, breakdown of rust resistance is emerging again, due to increased rust virulence associated to resistance genetic erosion during breeding.
The breakthrough idea of HDT-Coffee is to apply the unprecedented power of next generation sequencing (NGS) and computational analysis to implement targeted genomic approaches to characterize a unique set of HDT plants and derivatives hold by CIFC. The exceptional know-how in the coffee-rust interaction of CIFC, together with their exclusive plant and rust resources, will be combined with the top expertise in evolutionary biology and genomics of the CIBIO team, empowered by its NGS and computational platforms, to generate an in depth characterization of coffee rust resistance, and produce expedite molecular tools enabling to test the resistance status of present coffee crops, and guide marker-assisted gene pyramiding breeding for improved rust resistance. To achieve such goals, HDT-Coffee will perform high coverage sequencing of selected genomic regions from a collection of CIFC coffee plants including all rust resistance factors known in different combinations, and displaying an array of different behaviors upon inoculation with the full range of rust races isolated to date and conserved at CIFC. Advanced computational analysis will then harness the obtained genomic data to achieve three main objectives: decipher the contribution of HDT ancestors to the genome of their derivatives, identify and validate candidate loci explaining resistance phenotypes, and uncover the evolutionary history of resistance loci found in resistant and susceptible plants. The knowledge obtained will be used to generate two important deliverables: the development of a PCR-based protocol to identify the presence of resistance loci in coffee individuals, and the characterization of resistance profiles in worldwide cultivars, gathered by CIFC. HDT-Coffee will generate a deep understanding of the natural sources of rust resistance found in HDT coffee lineages, and will deliver key tools for the smart management of coffee crop resistance-status and breeding programs towards improved rust resistance. As such, HDT-Coffee will contribute to yield gains in the most economically important crop in the world, and to avoid massive antifungical treatments, lowering economical costs and environmental contamination. Upmost, HDT-Coffee should re-launch the central position of Portugal in the roadmap of coffee, as a provider of services and germplasm vital to the coffee industry. |
Exploiting genome wide sequence data for domestic species breeding (NEXT2)
AGL2016-78709-R (2017-2019)
Funded by the Ministery of Economy and Competitivity (MEC) / Ministery of Science and Innovation (MICINN)
PI: Sebastián E. Ramos Onsins & Miguel Pérez-Enciso
AGL2016-78709-R (2017-2019)
Funded by the Ministery of Economy and Competitivity (MEC) / Ministery of Science and Innovation (MICINN)
PI: Sebastián E. Ramos Onsins & Miguel Pérez-Enciso
|
Knowledge of the genetic architecture of complex traits and its dynamics is fundamental for understanding selective processes in domestic species. With this target in mind, here we propose studying the genetic architecture of complex traits at different time frames, from a few generations (genomic selection for commercially relevant traits) to thousands of generations (domestication), and its impact in practical breeding programs. Specifically, we propose:
To continue developing ngaSP software as an integrative computational platform, with the aim of making it a reference software in population genomics. We will include new functionalities, e.g., to visualize the effect of selection for a trait on genetic variation. To develop a tool that allows simulation of complex traits and complete genomes under flexible scenarios. To model the genetic architecture of complex traits considering pathway data and, in general, genome features information. To study the effect of the domestication on the whole genome. To combine statistical learning methods -useful to identify signals of association of the genotype with the phenotype- together with population genomics methodologies -searching for evolutionary models- in order to infer the effect of selection on the genome by means of the Site Frequency Spectra. Based on the previous results, to study the performance of genome selection using very high density marker data, up to imputed sequence and up to industry scale. To disentangle the haplotype structure of the porcine species, using the new assembly if available. To analyze the variability of genomic data from domesticated and wild animals (pig) and plants (cucurbitacea, rosacea) in order to understand the effects of the domestication on the genome. |
Next generation tools to exploit genome diversity in domestic species (NEXT)
AGL2013-41834-R (2014-2016)
Funded by the Ministery of Economy and Competitivity (MEC). PI: Sebastián E. Ramos Onsins & Miguel Pérez-Enciso.
AGL2013-41834-R (2014-2016)
Funded by the Ministery of Economy and Competitivity (MEC). PI: Sebastián E. Ramos Onsins & Miguel Pérez-Enciso.
|
The availability of complete genomes has dramatically improved the characterization of biodiversity and selection footprints in crop and livestock species, and has potentially many applications in practical breeding as well. Next Generation Sequencing (NGS) has certainly democratized genomics but, not unexpectedly, the dramatic increase in complete genome sequences has resulted in a bottleneck on how to optimally analyze these vast datasets. This project aims at providing methods and easy-to-use software (dnaSP) to analyze sequence data at the population level (Work Packages WP 1 and 2). The use of NGS data for genomic selection, in comparison to ascertained SNP arrays, will be also evaluated by simulation in several scenarios (WP 3). Finally, some porcine datasets of relevance to the industry and to the study of the effects of domestication and artificial selection will be analyzed (WP 4).
The dnaSP software will directly handle aligned bam files and will account for sequencing errors and incomplete data, typical features of NGS datasets. It will report classical and new statistics used in population genetics, e.g., to detect selective events. It will be programmed in a fully scalable manner such that new functionalities, e.g., association analyses or genomics selection could be added easily in the future. It will be publicly available. In WP3, A simulation study will be carried out to evaluate whether NGS would outperform dense genotyping and, if so, under what conditions. The effects of the demographic model, genotyping density vs. NGS coverage and QTL genetic architecture will be addressed. In WP4, several complementary analyses on a set of over 130 pig genomes available will be carried out to characterize polymorphism genome wide and species wide. We will study as well the genetic bases of domestication and of modern selection. These analyses will prompt us to utilize new concepts, in particular, we aim at incorporating functional information on the genome, such as pathway or metabolic databases. As a proof of concept, data from the industry of feed efficiency will be analyzed. |
Development of bioinformatic tools for the analysis of DNA variability at genomic scale. Application at domestic species
CGL2009-09346 (2010-2012)
Funded by the Ministery of Economy and Competitivity (MEC). PI: Sebastián E. Ramos Onsins
CGL2009-09346 (2010-2012)
Funded by the Ministery of Economy and Competitivity (MEC). PI: Sebastián E. Ramos Onsins
|
In the last years, the study of the nucleotide variation at the populations have been one of the main interests in many of biological fields. The knowledge of the levels and patterns of variation allow (between other aspects) to discriminate between selective and demographic processes, to detect regions of functional interest, to trace the history of the populations and to estimate the capacity of response of a population under specific selective events, in conjunction with association studies.
The proposal develop four main objectives: 1) To finish the development of a bioinformatic tool (with graphical interface) that allow to any researcher (expert or just interested in results from population genetics) the analysis of nucleotide variability of a small or massive quantity of experimental data. 2) To finish a second version of mlcoalsim, a tool for multilocus coalescent simulation analysis. This tool will be adapted to the study of large quantity of data (a large number of regions are large regions) with multiprocessor computation analysis. This tool will allow Bayesian analysis with the inclusion of prior distributions to almost all parameters. Furthermore, new demographic and selective models will be included, as well as recent speciation/isolation models. 3) Statistical analysis to determine the capability of the currrent methodologies that use summary statistics to detect demographic and selective events. 4) These tools and others will be used to the analysis of new generation sequencing data. Here, in collaboration with other collegues, a large number of DNA regions will be studied in domestic species (mainly porcine but also others). The analysis will be focus on finding the effect of selection in subdivided populations. It will be necessary the use of informatic equipment with medium/high calculation capacity. |